MetaADEDB: Introduction, Content and Use Cases
According to Food and Drug Administration (FDA), USA, Adverse Drug Reactions (ADRs) are one of the leading causes of morbidity and mortality in hospitals. Nearly 7000 estimated deaths occur per year due to ADRs in the USA. Although the pharmaceutical industry thoughtfully designs and conducts studies to develop a perfect drug with only targetted action and no adverse event, it is nearly impossible to have such a magic molecule. This impossibility arises due to the genetic, epigenetic, metagenomic, and environmental diversity of human beings. The factor that can significantly reduce the burden of ADRs on the patient and hospitals is the easy availability of known and reported side effects from published reports.
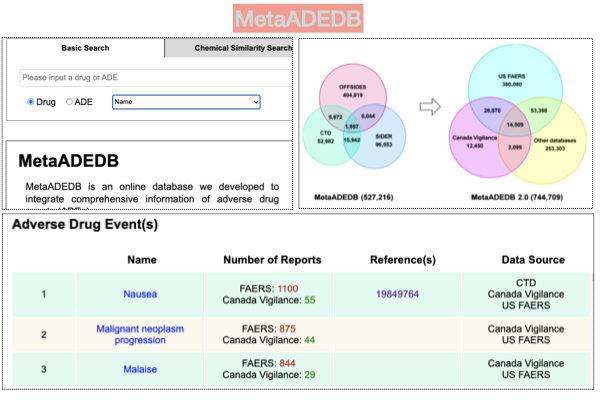
Several databases that compile and store adverse drug reactions have been developed and vary in quality and frequency of updates. A noticeable meta-database that aggregates and normalizes adverse drug reactions is a publicly accessible database: MetaADEDB. MetaADEDB aggregates information from the US FDA Adverse Event Reporting System (FAERS), Canada Vigilance Adverse Reaction, Comparative Toxicogenomics Database (CTD), Side Effect Resource (SIDER), and OFFSIDES.
The MetaADEDB was launched in 2013 and updated recently. Since 2013, researchers, family medicine physicians, and pharmaceutical personnel use this resource to access adverse drug reactions associated with drugs. Aggregated data about adverse drug reactions are supplemented with mapping to drug identifiers and phenotypes provided in PubChem, Drug Bank, and the Anatomical Therapeutic Chemical (ATC), UMLS, and MeSH. The current version of MetaADEDB has 744,709 drug-ADR associations between 8,498 drugs and 13,193 ADRs.
MetaADEDB supports searching using either name of the drug or identifier provided by authorities like PubChem, DrugBank, ChEMBL. Similarly, it supports name or identifier-based searches for adverse drug reactions. Other search modes include search based on chemical substructure or properties of molecules like molecular weight and LogP to support database mining for pharmaceutical personnel and researcher.
A typical results page includes information about the drug, basic molecular properties, and links to third-party resources. The result page lists adverse drug reactions and the number of patients developing them in the FAERS and Canada Vigilance database. For adverse drug reactions present in CTD and OFFSIDES, the result page links to PubMed articles.
Conclusions
MetaADEDB is a valuable resource that provides easy access to aggregated information for general practitioners and the pharmaceutical industry. Limitations of the database include the frequency of updates (updated only two times in the past eight years) and only provides basic information about the drug reactions.