Functional Evidence: A Comprehensive Guide for Variant Classification and Gene Validity Assessment
Introduction:
Genetic variant classification and establishing gene-disease associations are essential in clinical genetics and precision medicine. Functional assays, which assess the impact of genetic variants on gene and protein function, play a crucial role in these processes. Functional data provide insights into the pathogenicity of variants, aid in determining the underlying mechanisms of diseases, and guide therapeutic interventions. This article comprehensively reviews different types of functional assays and their use in variant classification and gene-validity assessments. We also discuss key considerations when using functional data to support variant classification.
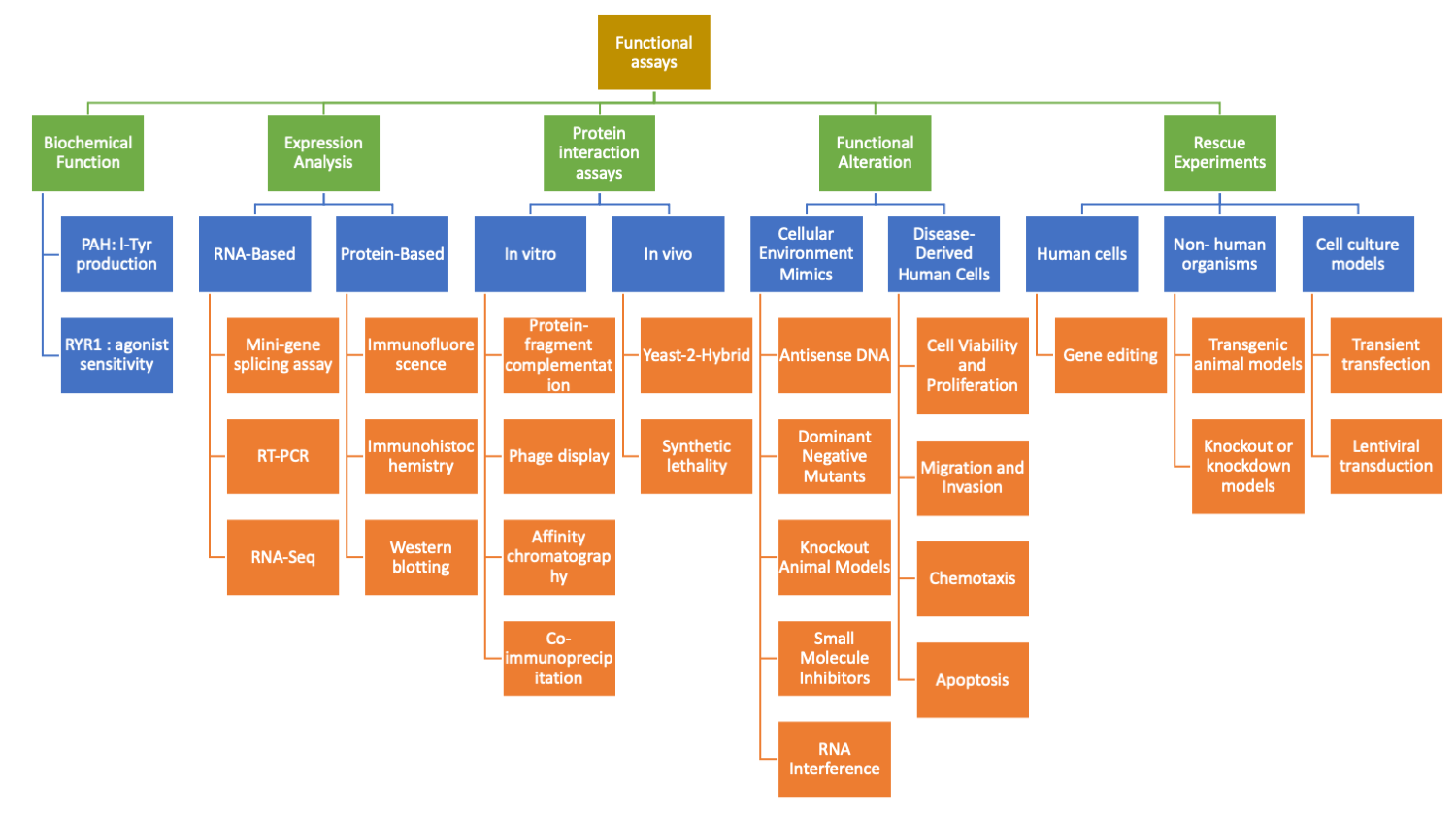
Biochemical Function:
One important category of functional data is a biochemical function, which evaluates the gene product's role in biological processes. Biochemical assays assess enzymatic activity in the presence of genetic variation. The impact of the variant on protein function can be determined by comparing the function of a variant with the wild-type gene product.
- GBA (Glucocerebrosidase)
- Enzymatic activity assay: Measurement of glucocerebrosidase activity using a fluorogenic substrate, such as 4-methylumbelliferyl-β-D-glucopyranoside (4-MUG). The cleavage of the substrate by the enzyme generates a fluorescent signal that can be quantified.
- Substrate degradation assay: Assessment of the ability of GBA to degrade its physiological substrate, glucocerebroside, by monitoring the reduction in substrate levels using mass spectrometry or high-performance liquid chromatography (HPLC).
- PTEN (Phosphatase and Tensin Homolog)
- Lipid phosphatase activity: Measurement of the phosphatase activity of PTEN using a synthetic phosphatidylinositol phosphate substrate, such as 3,3'-dihexadecylindocarbocyanine (DiI)-labeled phosphatidylinositol (3,4,5)-trisphosphate (PIP3). The dephosphorylation of PIP3 by PTEN can be quantified by monitoring the decrease in fluorescence intensity.
- PAH (Phenylalanine Hydroxylase)
- Quantification of l-Tyr production: Assessment of the enzymatic activity of PAH by measuring the production of l-tyrosine from phenylalanine using various methods such as high-performance liquid chromatography (HPLC), spectrophotometry, or fluorescent probes.
- RYR1 (Ryanodine Receptor 1)
- Increased sensitivity to RYR1 agonist in HEK293 cells: Functional assessment of RYR1 variant impact on calcium release by measuring intracellular calcium levels in HEK293 cells expressing the wild-type or variant RYR1. The cells are stimulated with a specific RYR1 agonist, such as 4-chloro-m-cresol (4-CmC), and the resulting calcium release can be quantified using calcium-sensitive dyes or fluorescence microscopy.
These assays provide insights into the biochemical functions of GBA, PTEN, PAH, and RYR1, helping to determine the functional consequences of genetic variants and their impact on protein function and cellular processes.
Expression Analysis:
Expression analysis provides valuable insights into gene expression levels at the RNA and protein levels, which can help assess the functional impact of genetic variants. Here, we will discuss two expression analysis categories: RNA-based and protein-based.
RNA-Based Expression Analysis:
- RNA-Seq (RNA Sequencing): RNA-Seq is a powerful, comprehensive gene expression analysis technique. It involves sequencing RNA molecules, providing information about the abundance and diversity of transcripts in a sample. RNA-Seq can help identify differential expression patterns between different samples, including diseased and normal tissues, providing insights into the dysregulation of gene expression associated with genetic variants.
- RT-PCR (Reverse Transcription Polymerase Chain Reaction): RT-PCR is a widely used technique to measure the levels of specific RNA transcripts. It involves reverse transcription of RNA into complementary DNA (cDNA), followed by PCR amplification of the target transcript. RT-PCR can provide quantitative information about the expression levels of specific genes, allowing for comparing gene expression between different samples or conditions.
- Mini-gene splicing assay: This assay involves the construction of a synthetic gene fragment containing the exon(s) of interest and flanking intronic sequences. The mini-gene is transfected into cells, and the splicing pattern is analyzed using RT-PCR. Introducing specific variants into the mini-gene makes it possible to assess their impact on splicing. Aberrant splicing patterns observed in the presence of variants can indicate their potential pathogenicity.
Protein-Based Expression Analysis:
- Western blotting: Western blotting is widely used to detect and quantify specific proteins in a sample. It involves protein extraction, gel electrophoresis separation, membrane transfer, and subsequent antibody-based detection. Western blotting allows for the assessment of protein expression levels and can be used to compare protein levels between different samples or conditions.
- Immunohistochemistry (IHC): IHC is a technique that enables the visualization and localization of specific proteins within tissue sections. It involves using antibodies labeled with enzymes or fluorescent dyes to detect the presence of target proteins. IHC can provide information about the spatial distribution and relative abundance of proteins in tissues, helping to elucidate their role in disease processes.
- Immunofluorescence (IF): Like IHC, immunofluorescence utilizes fluorescently labeled antibodies to detect specific proteins. However, instead of using enzyme-based detection, IF relies on the detection of fluorescence signals. This technique allows for the visualization of protein expression patterns within cells or tissues, providing insights into protein localization and cellular processes.
These expression analysis techniques, both at the RNA and protein levels, offer valuable information about gene expression patterns, protein levels, and subcellular localization. By comparing expression profiles between diseased and normal samples or assessing the impact of genetic variants on gene expression or protein levels, these assays contribute to our understanding of the functional consequences of genetic variants and disease association.
Protein interaction assays
Protein-protein interactions play a crucial role in various biological processes, and alterations in these interactions can have significant implications for disease development. Assessing protein-protein interactions can provide valuable insights into the functional impact of genetic variants and help in the classification of variants associated with diseases. Two broad categories of protein-protein interaction assays include in vitro assays and in vivo assays.
In vitro protein-protein interaction assays:
In vitro assays are performed outside of a living organism and provide controlled experimental conditions to study specific protein-protein interactions. Here are some examples:
- Co-immunoprecipitation (coIP): CoIP is a widely used assay that involves the immunoprecipitation of a target protein along with its interacting partners from a cellular lysate. The coIP assay can confirm physical interactions between proteins and can help assess the impact of variants on these interactions. For example, in the context of breast cancer-associated genes BRCA1 and BRCA2, coIP assays have been used to evaluate the interaction between these proteins and key binding partners like RAD51 and PALB2.
- Affinity chromatography: Affinity chromatography involves the immobilization of a target protein on a solid support matrix, followed by the isolation of its interacting partners using specific affinity tags or antibodies. This technique allows the identification and characterization of protein-protein interactions and can be used to study the impact of variants on these interactions.
- Protein-fragment complementation: Protein-fragment complementation assays utilize split reporter protein systems where the complementation of two protein fragments occurs upon the interaction between two target proteins. The reconstitution of the functional reporter protein indicates protein-protein interaction. This assay can be employed to study the disruption of protein-protein interactions caused by genetic variants.
- Phage display: Phage display is a powerful technique that utilizes bacteriophages to express and present proteins on their surfaces. By constructing libraries of peptide or protein variants, interactions with target proteins can be assessed. Phage display has been used to identify protein-protein interactions and evaluate the impact of genetic variants on these interactions.
In vivo protein-protein interaction assays:
In vivo assays examine protein-protein interactions within a living organism, providing insights into the physiological relevance of these interactions. Here are a couple of examples:
- Yeast-2-Hybrid (Y2H): Y2H is a commonly used in vivo assay performed in yeast cells to identify and validate protein-protein interactions. By fusing target proteins to different domains of a transcription factor, protein interactions can be detected through reconstitution of the transcription factor and subsequent reporter gene activation. Y2H has been employed to study protein-protein interactions in various disease-related genes and assess the impact of variants on these interactions.
- Synthetic lethality assays: Synthetic lethality occurs when the combined disruption of two genes leads to cell death, while disruption of either gene alone does not. Synthetic lethality assays can be used to identify genetic interactions and assess the functional consequences of genetic variants. By evaluating the impact of variants on synthetic lethality interactions, insights can be gained into the role of specific variants in disease development.
Examples of protein-protein interaction assay in variant classification:
- BRCA1/BRCA2: Genetic variations in BRCA1 and BRCA2 predisposes an individual to hereditary breast and ovarian cancer syndrome (HBOC). Protein-protein interaction assays, such as coIP and Y2H, have been used to investigate the interactions between BRCA1 and BRCA2 with key binding partners like RAD51 and PALB2. These assays help evaluate the impact of variants on these interactions and provide evidence for variant classification.
- TP53: Variants in the TP53 gene are associated with Li-Fraumeni syndrome and other cancers. Protein-protein interaction studies, including coIP and affinity chromatography, have been utilized to assess the impact of TP53 variants on interactions with other proteins involved in the p53 pathway, such as MDM2 and p300. These interactions provide insights into the functional consequences of TP53 variants and aid in variant classification.
- These examples highlight how protein-protein interaction assays contribute to the understanding of variant effects and their relevance to disease. By studying the impact of variants on protein-protein interactions, researchers can unravel the molecular mechanisms underlying genetic diseases and inform variant classification decisions.
Functional Alteration:
Functional alteration studies are essential for understanding how genetic variants impact gene function and contribute to disease. In this section, we will discuss two categories of functional alteration studies: 1) Disease-derived human cells and 2) Assays using cellular environment mimics.
Disease-Derived Human Cells:
Disease-derived human cells are a valuable resource for studying the functional consequences of genetic variants in a relevant biological context. These cells are derived from patients with the specific disease of interest and can provide insights into how genetic variants affect cellular phenotypes. Various functional assays can be performed using disease-derived human cells to assess the impact of genetic variants, including:
- Cell Viability and Proliferation Assays: These assays measure the growth and viability of cells with specific genetic variants compared to control cells. Techniques such as MTT assay, ATP measurement, or cell counting can be employed to evaluate cell viability and proliferation rates.
- Migration and Invasion Assays: These assays assess the migratory and invasive capabilities of cells harboring genetic variants. Techniques like transwell migration assay or Boyden chamber assay are used to measure the cell's ability to migrate and/or invade through a porous membrane.
- Chemotaxis Assays: Chemotaxis assays evaluate the ability of cells to respond to chemical gradients. They provide insights into the role of specific genetic variants in directing cell movement towards or away from particular signals.
- Apoptosis Assays: Apoptosis assays measure the programmed cell death response in cells with genetic variants. Techniques such as Annexin V staining or TUNEL assay can be used to detect apoptotic cells and evaluate the impact of variants on cell survival.
Assays Using Cellular Environment Mimics:
- Assays utilizing cellular environment mimics aim to recreate specific cellular conditions or perturbations to understand gene function and the impact of genetic variants. These assays can provide insights into the role of particular genes in disease processes and help elucidate potential therapeutic targets. Some commonly used methods in this category include:
- RNA Interference (RNAi): RNAi involves the introduction of small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs) to specifically knock down the expression of target genes. By observing the phenotypic changes resulting from gene knockdown, the functional consequences of genetic variants can be assessed.
- Antisense DNA: Antisense DNA technology utilizes synthetic DNA molecules that are complementary to specific target gene sequences. By targeting and binding to mRNA molecules, antisense DNA can modulate gene expression and provide insights into the functional impact of genetic variants.
- Dominant Negative Mutants: Dominant negative mutants are altered forms of proteins that interfere with the normal function of the wild-type protein. By expressing dominant negative mutants, the impact of genetic variants on protein function can be evaluated, particularly in cases where the variant disrupts protein-protein interactions or enzymatic activity.
- Knockout Animal Models: Knockout animal models involve the generation of genetically modified animals lacking the expression of a specific gene. These models allow for the study of gene function in a living organism and the evaluation of the consequences of genetic variants on physiological processes.
- Small Molecule Inhibitors: Small molecule inhibitors are compounds that target and inhibit the activity of proteins or enzymatic pathways. By treating cells or animal models with small molecule inhibitors, the functional consequences of genetic variants can be assessed by observing the phenotypic changes resulting from pathway inhibition.
Rescue Experiments:
Rescue experiments play a crucial role in understanding the functional consequences of genetic variants. Here, I will summarize the various approaches and model systems used for phenotype rescue and provide examples of successful rescues:
Rescue in human cells:
- Introduction of wild-type gene or gene product: Patient-derived cells with a genetic defect can be transfected or transduced with exogenous wild-type genes or gene products. Successful rescue of the phenotype indicates that the variant gene is responsible for the observed functional defect. Examples include the rescue of Zellweger-spectrum disorders by transfection of patient-derived fibroblasts with a combination of wild-type peroxisomal gene and peroxisomally targeted green fluorescence protein (GFP).
- Gene editing: Techniques such as CRISPR-Cas9 are employed to precisely edit the variant gene in patient cells, restoring the wild-type sequence or function. This approach has been used in studies of mitochondrial disorders, where lentiviral transduction of patient fibroblasts corrected the deficiency of respiratory chain enzymes.
Rescue in non-human model organisms:
- Transgenic animal models: Animal models with the variant gene can be generated, and rescue experiments involve introducing the wild-type gene or its functional equivalent. Successful restoration of normal physiological parameters, behavior, or disease-specific manifestations demonstrates rescue. For example, transgenic mouse models have been used to rescue phenotypes associated with genetic variants in various genes.
- Knockout or knockdown models: In these models, the variant gene is overexpressed or knocked out, respectively, and the rescue is achieved by introducing the wild-type gene or its product. Restoration of normal phenotypes indicates successful rescue.
Rescue in cell culture models:
- Transient transfection: Patient-derived cells or model systems expressing the variant gene can be transfected with exogenous wild-type genes. The rescue is assessed by functional assays specific to the gene of interest. For example, rescue experiments using transient transfection have been conducted in yeast, where the growth on fermentable and non-fermentable carbon sources is assessed to study mitochondrial disorders.
- Lentiviral transduction: Patient-derived fibroblasts or iPSCs can be transduced with lentiviral vectors carrying the wild-type gene. The rescue is evaluated by measuring functional or biochemical parameters. Lentiviral transduction has been successfully used in studies of mitochondrial disorders, including oxidative phosphorylation deficiencies.
- It is essential to consider proper controls and carefully interpret the results of rescue experiments. Controls should include negative controls for the transduction or transfection procedure, such as the transduction of patient cells with the mutant gene as an additional negative control. Furthermore, competition between endogenous and exogenous gene products should be taken into account.
In summary, rescue experiments utilizing different model systems, including human cells, non-human model organisms, and cell culture models, provide valuable evidence of the functional consequences of genetic variants. Successful rescues by introducing wild-type genes or gene products demonstrate the role of specific variants in the observed phenotypes and contribute to variant classification and understanding of disease mechanisms.
Considerations and caveats in using functional data in gene-validity assessments and variant classification
When interpreting functional data for variant classification, it is essential to consider several considerations and caveats. Here are some critical points to keep in mind:
- Contextual relevance: Functional data should be interpreted in the context of the gene's normal function, the specific tissue or cell type affected, and the overall disease pathology. Not all genes or variants have well-established functional assays, and the relevance of functional studies may vary depending on the gene's role in a particular biological pathway or disease mechanism.
- Assay limitations: Functional assays have inherent limitations, including sensitivity, specificity, and reproducibility. Different assays may provide complementary information, but no single assay can capture the complete complexity of gene function. It is crucial to understand the strengths and limitations of each assay used to assess functional impact.
- Model systems: The choice of a model system for functional studies is critical. Different model systems (e.g., human cells, non-human model organisms, cell culture models) have unique advantages and disadvantages. It is important to consider the relevance of the model system to the human disease being studied and to carefully interpret and extrapolate findings from one system to another.
- Phenotype-genotype correlation: The relationship between genotype and phenotype can be complex, and functional studies may not always correlate directly with clinical presentation. Variants can have variable penetrance, expressivity, and modifying factors, making it challenging to predict the clinical consequences based solely on functional data.
- Genetic background: Genetic modifiers, including variants in other genes, can influence the phenotypic outcome of a specific variant. Functional studies may not always capture the interplay between multiple genetic factors, and the specific genetic background should be considered when interpreting functional data.
- Complementary evidence: Functional data should be evaluated alongside other lines of evidence, such as population frequency, segregation data, computational predictions, conservation, and clinical data. Integrating multiple lines of evidence strengthens the overall assessment of variant pathogenicity.
- Technical artifacts: It is important to account for potential technical artifacts that may arise during experimental procedures, such as non-specific binding, off-target effects, or unintended consequences of genetic manipulation techniques. Appropriate controls and replication of experiments are crucial to validate functional findings.
- Variant interpretation: Functional data should contribute to a comprehensive assessment of variant pathogenicity but should not be considered in isolation. Functional data, along with other evidence, should be weighed collectively to determine the overall classification of a variant according to established variant classification guidelines.
Briefly, while functional data provide valuable insights into the impact of genetic variants, their interpretation requires careful consideration of contextual relevance, assay limitations, model systems, phenotype-genotype correlations, genetic background, complementary evidence, potential artifacts, and integration with other lines of evidence.
Conclusion:
Functional data provide valuable insights into the impact of genetic variants on gene and protein function, aiding in variant classification and gene-validity assessments. Assays assessing biochemical function, expression analysis, functional alteration, model systems, rescue experiments, and massively parallel functional assays contribute to our understanding of gene-disease associations. However, careful consideration of physiological context, molecular consequences, assay performance, and appropriate controls is essential for accurate interpretation. By incorporating these considerations, functional data can enhance the classification of genetic variants and provide important insights into disease mechanisms and potential therapeutic interventions.
References
- The functional genomics laboratory: functional validation of genetic variants PMC
- Gene Clinical Validity Curation Process: Standard Operating Procedure Version 7.0 ClinGen
- Variant Interpretation: Functional Assays to the Rescue. PMC
- Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Article
- Refinement of the assignment to the ACMG/AMP BS3 and PS3 criteria of eight BRCA1 variants of uncertain significance by integrating available functional data with protein interaction assays. Article
- Comparative analysis of functional assay evidence use by ClinGen Variant Curation Expert Panels. Article
- Classification of MSH6 Variants of Uncertain Significance Using Functional Assays. Article